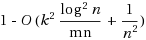Combinatorial (or rule‐based) methods for inferring haplotypes from genotypes on a pedigree have been studied extensively in the recent literature. These methods generally try to reconstruct the haplotypes of each individual so that the total number of recombinants is minimized in the pedigree. The problem is NP‐hard, although it is known that the number of recombinants in a practical dataset is usually very small. In this paper, we consider the question of how to efficiently infer haplotypes on a large pedigree when the number of recombinants is bounded by a small constant, i.e. the so called k‐recombinant haplotype configuration (k‐RHC) problem. We introduce a simple probabilistic model for k‐RHC where the prior haplotype probability of a founder and the haplotype transmission probability from a parent to a child are all assumed to follow the uniform distribution and k random recombination events are assumed to have taken place uniformly and independently in the pedigree. We present an O(mnlog
k+1
n) time algorithm for k‐RHC on tree pedigrees without mating loops, where m is the number of loci and n is the size of the input pedigree, and prove that when 90 log n
<m
<n
3, the algorithm can correctly find a feasible haplotype configuration that obeys the Mendelian law of inheritance and requires no more than k recombinants with probability
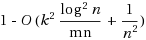 . The algorithm is efficient when k is of a moderate value and could thus be used to infer haplotypes from genotypes on large tree pedigrees efficiently in practice. We have implemented the algorithm as a C++ program named Tree‐k‐RHC. The implementation incorporates several ideas for dealing with missing data and data with a large number of recombinants effectively. Our experimental results on both simulated and real datasets show that Tree‐k‐RHC can reconstruct haplotypes with a high accuracy and is much faster than the best combinatorial method in the literature.
. The algorithm is efficient when k is of a moderate value and could thus be used to infer haplotypes from genotypes on large tree pedigrees efficiently in practice. We have implemented the algorithm as a C++ program named Tree‐k‐RHC. The implementation incorporates several ideas for dealing with missing data and data with a large number of recombinants effectively. Our experimental results on both simulated and real datasets show that Tree‐k‐RHC can reconstruct haplotypes with a high accuracy and is much faster than the best combinatorial method in the literature.